Archive
eMarch 4: Compilation environment variable

I sometime use improved library like mkl, in my code, the later need to have their path added to the LD_LIBRARY_PATH, in order to be accessed during compilation. Eventually, this may prevent the very handful M-x compile command to work with emacs. The fix I found on stackoverflow
(let ((path (shell-command-to-string ". ~/.bashrc; echo -n $PATH"))) (setenv "PATH" path) (setq exec-path (append (split-string-and-unquote path ":") exec-path))) (let ((ld_library_path (shell-command-to-string ". ~/.bashrc; echo -n $LD_LIBRARY_PATH"))) (setenv "LD_LIBRARY_PATH" ld_library_path) (setq exec-path (append (split-string-and-unquote ld_library_path ":") exec-path)))
Glascow
 After some months of work, we had our article accepted in bioinformatics and the accompanying software GLASCOW released (available on the GIGA website).
After some months of work, we had our article accepted in bioinformatics and the accompanying software GLASCOW released (available on the GIGA website).
Basically, the software perform a generalized linear mixed model analysis on (preferably) hidden states (obtained e.g. with PHASEBOOK) after a correction for population stratification. This approach has already been successfully applied in : Durkin et al, Sartelet et al, Dupuis et al.
I contribute to the software with some optimizations and code rewriting. The code is thus a mix of several coding practice and style, I must admit that to some extent this can made the code hard to follow.
Seen on this blog
Concerning programming, several details of the package are available on the blog :
- The general framework of the package was set by the script LancementProjet.sh
- I use command argument as described here
- The use of Lapack is conditional, and therefore I add some preprocessor instructions in the code
- In order to get info on date of compilation I also add some preprocessor instructions
…coming soon on this blog ?
Others key features are :
- A parallelized reading of the files (we rely on the use of one phase/hidden states file per chromosome, obtained with PHASEBOOK), this trick plus a neat storage of the partial relationship between individuals allow to obtain great speed improvements and lower memory consumption.
- The computation of score on residuals allow both a general speed-up and an ability to restart analysis only from these last steps.
- Permutations are done in parallel, nevertheless this last part was very tricky to implement, and I must says that the speed-up didn’t fulfill my expectations….I’ll try to see how to fix this.
- We add an option for outputting results in a “.assoc” file for a better viewing experience in IGV.
So even if the code still contain some old fashion Fortran (goto etc…) the adventure was fun, inspired me some posts, give me some experience in glmm and add one article to my publications list ! Not so bad, isn’t it ?
Tips : Reading compressed file with fortran and named pipe
 As genomic data are accumulating at some point we have to find solutions to reduce data size as much as possible. Working with beagle (java software), made me noticing that most of “modern language” have standard library to read directly compressed file. Alas most of my tools are written in fortran and the few example I’ve seen of direct reading of compressed files used socket or bindings to C system calls….which are far from the ease of use of python or java library.
As genomic data are accumulating at some point we have to find solutions to reduce data size as much as possible. Working with beagle (java software), made me noticing that most of “modern language” have standard library to read directly compressed file. Alas most of my tools are written in fortran and the few example I’ve seen of direct reading of compressed files used socket or bindings to C system calls….which are far from the ease of use of python or java library.
Fortunately, I discover recently the named pipe. The idea is to create a “file” that will behave like a pipe. One process will read whatever enter in the pipe and another will write in it.
The advantage here are :
- You don’t have to make changes in your fortran program
- You don’t need to decompress entirely a file to read it (and therefore you spare some disk space).
One example :
You have a compressed file containing genotypes. You need to read the genotype from your program, but you don’t have enough space available to decompress entirely your file. The idea is then to create a named pipe.
Named pipe creation
Named pipe can be created with the following command :
mkfifo MyNamedPipe
A simple ls -al should confirm the creation of the named pipe with the following line :
prw-rw-r– 1 francois francois 0 2012-01-07 19:53 MyNamedPipe
Redirections to named pipe
Suppose you want to compress directly a big file that you generate with any (let say fortran) program. We’ll proceed following these steps :
- In a first terminal, start the compression process on the pipe file (e.g. gzip MyNamedPipe)
- In a second terminal, write to the named pipe some data
Example with fortran
program testfifo implicit none integer :: i,j open(11,file="MyNamedPipe") do i=1,10000 do j=i,10000 write(11,*)i,j,j*i end do end do end program testfifo
Compilation
gfortran -o testfifo testfifo.f90
Execution
./testfifo
=> As the named pipe is empty the program is waiting for data
In another shell :
cat MyNamedPipe | gzip >T.gz
And then voilà the decompression start and by turn fortran program continue its normal execution.
Fortran
Things are working, but it would be nice to use mkfifo more smoothly. This can be achieve with the EXECUTE_COMMAND_LINE subroutine, within a fortran program.
Two problems should be fixed then :
- We should not wait the call system to finish in order to start the reading process.
- The read loop should know when file is totally decompressed (in fact as any other stream could be redirected to the named pipe, no end of file signal will be received by the loop).
The first point is obtained by the “&” sign to put the process in background. The second point, is obtained by echoing the “end of file” sign to the named pipe before removing it. So basically, we end-up with something like the following code.
program testfifo
implicit none
integer:: i,j,k,io,unit,iostat
character(len=30)::File,filename
character(len=600)::Instruction
write(*,*)Name of the xz file ?"
read(*,*)filename
!//Create a named pipe//
call EXECUTE_COMMAND_LINE("rm -f MyUnCompressedFile ;mkfifo MyUnCompressedFile")
!//Open a connection to it//
open(11,file="MyUnCompressedFile",iostat=iostat)
!//Write decompression instruction//
write(Instruction,'(a,a,a)')"(xz -dc ",filename," >MyUnCompressedFile; echo \x4 >MyUnCompressedFile )"
!//Execute//
call EXECUTE_COMMAND_LINE(Instruction)
!//Execute a normal reading loop//
do
read(11,*,iostat=io)i,j,k
if(io/=0)exit
write(*,*)i,j,k
end do
!//remove named pipe//
write(Instruction,'(a)')"( rm -f MyUnCompressedFile)"
CALL EXECUTE_COMMAND_LINE(Instruction)
end program testfifo
Compilation
gfortran -o testfifo testfifo.f90
Execution
echo T1.xz | ./testfifo
Note : We compile the program with gfortran. In fact, ifort doesn’t recognize (for the moment), the EXECUTE_COMMAND_LINE subroutine, which is part of the new fortran 2008 standard. In order to have the following program working, you should use “call system” instead of “call EXECUTE_COMMAND_LINE” (the first beeing an extension to fortran recognized by both ifort and gfortran).
One Last step beyond
In order to get an even smoother way to read compressed data, we can write a module with function that will work as open and read….you can find here such a module….by the way the reading part work fine, I still haven’t found a good way to open a named pipe for output compression (any hints would be very appreciated !)
Using Fortran preprocessor (2)
Following last post on Fortran and preprocessing, we ‘ll try to see other use of preprocessing instructions.
Fortran accept several preprocessor instructions, we’ve illustrated the conditional preprocessor’s instructions, let’s look now at some case studies and how preprocessor’s instructions can help.
Debugging
Suppose you are given a very buggy code, (I presume that you would not dare writing bugs by your own :-p) your compiler can’t help you to trace the origin of the error (which is obviously a naïve statement), so you decide to start a long and boring task consisting in adding through out the code some tags that will tell you were are you in the code…the normal way is to write several time a line like :
write(*,*)"program reach the point A" .... write(*,*)"program reach the point B"
And then you ‘ll have to find where is A or B, to identify where the “first” bugs happens.
Thanks to the “__LINE__” instruction, you’ll be able to retrieve the line of code executed. So our instructions will be something like this
write(*,*)"program reach the line",__LINE__
Note, that the “_” is doubled before and after the LINE keyword.
Let’s assume your source code is split in several files, so to avoid any confusion, we’ll use another instruction
“__FILE__“, which, as you’ve probably guess, contain the name of the file containing the code that is being executed.
We now have something like :
write(*,*)"program reach the line",__LINE__," in file ",__FILE__
And now the “hammer”, insert this line in your code with someting like :
for FIC in *.F90
do
mv $FIC ${FIC}.bck
gawk '{if((NR%10)==0){print $0 "\n write(*,*)\"program reach the line\",__LINE__,\" in file \",__FILE__"}else{print $0}}' ${FIC}.bck >$FIC
done
Compile and test !
NB : The \” symbols are there because otherwise ” would be interpreted by awk as the end of a character string.
Once, you’ve corrected all the bugs, turn things back to normal with let’s say… grep :
for FIC in *F90
do
mv $FIC ${FIC}.dbg
grep -v "program reach the line" ${FIC}.dbg >>${FIC}
done
Avoiding debugging
Most often, when people report a bug, they never mention where does the binary they used come from, this, of course, can very often explain why the binary doesn’t make the new feature describe in your manual (at least the user read the manual !), but even with some indications on the software version, it is not always easy to know when the code has been compiled (and if you’ve eventually change your makefile as instance).
So of course, here I’ll present both __DATE__ and __TIME__ instructions, the latter giving the date of compilation and the time (in conjunction with a makefile under version control, you should be able to know everything about how your binary was compiled).
A nice way to use these instructions is by mean of command arguments, with a flag “–version” as instance.
so the example given in the command line post would then be :
...
if(narg>0)then
!loop across options
do cptArg=1,narg
call get_command_argument(cptArg,name)
select case(adjustl(name))
case("--version","-v")
write(*,*)"This is program TestArg : Version 0.1"
write(*,*)"Binary compiled the",__DATE__," at ",__TIME__
stop
case default
write(*,*)"Option ",adjustl(name),"unknown"
end select
end do
end if
...
Note for people using svn as instance, you can also add an $Id: $ flag in your code so that you ‘ll then know both the revision ID and the compilation date.
...
if(narg>0)then
!loop across options
do cptArg=1,narg
call get_command_argument(cptArg,name)
select case(adjustl(name))
case("--version","-v")
write(*,*)"This is program TestArg : Version $Id: $"
write(*,*)"Binary compiled the",__DATE__," at ",__TIME__
stop
case default
write(*,*)"Option ",adjustl(name),"unknown"
end select
end do
end if
...

So now, you’ve seen two possible uses of preprocessor’s instructions, we’ll see in the next post of the serie some other use of preprocessor.
Using Fortran preprocessor (1)
 Library like BLAS or LAPACK are both very efficient and pretty often installed on linux computer. Anyway, when you have to share a source code, you can’t rely on the fact that those libraries are set up on the host computer. So the safe solution is to find a potentially less efficient alternative, which may sound annoying for people used to BLAS and LAPACK.
Library like BLAS or LAPACK are both very efficient and pretty often installed on linux computer. Anyway, when you have to share a source code, you can’t rely on the fact that those libraries are set up on the host computer. So the safe solution is to find a potentially less efficient alternative, which may sound annoying for people used to BLAS and LAPACK.
Fortunately, for this kind of dilemma there is the PreProcessor, that although not part of the standard, is understood by several fortran compiler (at least gfortran, ifort and xlf).
The basic idea of preprocessing is to allow conditional compilation : depending on certain conditions, one part or another of the code will be compiled.
A first exemple
To illustrate let’s make a sample program, that should be named “TestPreProcessor.F90”, the extension should be written with capital letters (i.e .F90 and not .f90). That’s the way most compilers automatically know that they should preprocess the code.
program TestPreProcessor implicit none #if TEST write(*,*)"Compiled with Test Flag" #ELIF write(*,*)"Not Compiled with Test Flag" #END end program TestPreProcessor
Now we have two ways to compile our program.
The classical way
#Compile with no flag ifort TestPreProcessor.F90 -o TestPreProcessor #Test ./TestPreProcessor #You should see something like this: Not Compiled with Test Flag
Now the alternative way
#Compile with no flag ifort -DTEST TestPreProcessor.F90 -o TestPreProcessor #Test ./TestPreProcessor #You should see something like this: Compiled with Test Flag
The important point is that the flag -D stand for “define”, then during preprocessing, the keyword written after -D will be considered as defined. The keys #IF…#ELIF…#END will then be examined.”#if TEST” will stand for “is TEST defined ?” if it is true then code below the #if will compiled otherwise code above #elif will be compiled. That ‘s as simple as it seems !
Wrap up and useful example
So we ‘ve explained the basic idea with preprocessing, let apply this to a real problem. Suppose, we want to make a dot product a two vectors, and would like to offer the possibility to use either the dot_product intrinsic or the equivalent lapack subroutine.
program Dotprod implicit none integer,parameter::n=1000000 real(kind=4)::V1(n),V2(n),results real,external::sdot V1=1.0; V2=1.0 !//Use BLAS if asked// #if BLAS results=sdot(n,V1,1,V2,1) #else results=dot_product(V1,V2) #endif !//only a test// write(*,*)results end program Dotprod
And the corresponding makefile could look like :
PROG=Dotprod COMP=ifort #-------------------------------------------------------------------- #Compilations options + Library path #-------------------------------------------------------------------- BLAS= -DBLAS -lblas FF= -O3 $(BLAS) # Comment or uncomment $(BLAS) #-------------------------------------------------------------------- #Main rules #-------------------------------------------------------------------- $(PROG): $(PROG).F90 $(COMP) $(FF) -o $(PROG) $(PROG).F90
 These should allow to use either a BLAS subroutine or a standard internal subroutine throughout your code, and ask for very few changes to the end-user who would like to compile your code.
These should allow to use either a BLAS subroutine or a standard internal subroutine throughout your code, and ask for very few changes to the end-user who would like to compile your code.
There are several other very useful preprocessor reserved key…but we will see them in an other post 🙂
Fortran command line arguments
![]() One little detail that can make your program looks professional is
One little detail that can make your program looks professional is
the ability to pass arguments on the command line.
Surprisingly a lot of Fortran programmers don’t know that this can be done easily, this post will give some tips to use commands arguments.
First, until recently Fortran standard didn’t include any instructions to handle command line argument. Nevertheless, several compilers (in fact all the compilers, I’ve worked with) like ifort, gfortran, g95 were accepting the use of C function iargc and the subroutine getarg. And I must confess that until recently this was the tools I used.
Anyways since more and more fortran compilers are compliant with the F03 standard, the official (and recommended) way to deal with command line arguments is the function :
command_argument_count()
which retrieve the number of argument passed to the command line
and the subroutine
get_command_argument(i,aname)
which put character string present at the “i”th argument into variable “aname”.
First step
To illustrate use of command arguments let’s make a sample program
program TestArg
integer::narg,cptArg !#of arg & counter of arg
character(len=20)::name !Arg name
!Check if any arguments are found
narg=command_argument_count()
!Loop over the arguments
if(narg>0)then
!loop across options
do cptArg=1,narg
call get_command_argument(cptArg,name)
select case(adjustl(name))
case("--help","-h")
write(*,*)"This is program TestArg : Version 0.1"
case default
write(*,*)"Option ",adjustl(name),"unknown"
end select
end do
end if
end program TestArg
Compile and test.
ifort TestArg.f90 -o TestArg ; ./TestArg --help
This is for the basic case. A good hint is trying to use “classical” flags, in fact many software use somehow the same flag for classical purpose “verbosity”, “help” etc… I do believe that the arguments are really helpful because they allow you to offer a variety of options that won’t bother the user by default. In fact, without optional arguments, we tend to ask the user a lot of information that are for common users either obvious or useless.
A convenient use of the command arguments, is to create a debug flag that will help to correct your program by enabling the printing of more information, or version flag that can tell you which version of the software are you using…these flags can reduce tremendously your debugging time !
Adding some difficulties….
I generally use arg to activate an option, and later in the program I execute some pieces of code to be executed. As instance, if I want to write down a certain file to be read, I add a flag and later in the program execution, I will ask for the name of this file. Anyway, you probably know some software (plink, beagle as instance) which looks for all the information they need in the command arguments : With plink, you would as instance write :
plink --ped mydata.ped --map autosomal.map
A program such as the previous one, won’t handle such situation…but be can program something similar (although I believe this is not so nice).
program TestComplexArg
integer::narg,cptArg
character(len=20)::name,pedFile,mapFile
logical::lookForPed=.FALSE.
logical::lookForMap=.FALSE.
logical::fileExist
!Check if arguments are found
narg=command_argument_count()
if(narg>0)then
!loop across options
do cptArg=1,narg
call get_command_argument(cptArg,name)
select case(adjustl(name))
!First known args
case("--ped")
lookForPed=.TRUE. !change logical value
case("--map")
lookForMap=.TRUE.
case default
!Treat the second arg of a serie
if(LookForPed)then
pedFile=adjustl(name) !assign a value to pedfile
inquire(file=pedFile,exist=fileExist)!check if it exist
if(.not.fileExist)then
write(*,*)'file ',pedFile,' not found'
stop
endif
LookForPed=.FALSE. !put the logical variable to its initial value
elseif(LookForMap)then
mapFile=adjustl(name)
inquire(file=mapFile,exist=fileExist)
if(.not.fileExist)then
write(*,*)'file ',mapFile,' not found'
stop
endif
LookForMap=.FALSE.
else
write(*,*)"Option ",adjustl(name),"unknown"
endif
end select
end do
endif
end program TestComplexArg
Compile and test.
#Compile ifort TestArg.f90 -o TestArg #Create empty files touch mydata.ped autosomal.map #Test ./TestArg --ped mydata.ped --map autosomal.map
I hope these examples can be useful, and allow you to code “better” (either less verbose or more user-friendly) software.
Ifort Quick tips
Ifort is a pretty nice fortran compiler, nevertheless one thing that I find a little bit annoying is its default record length format, when you are testing /debugging a program, it’s always convenient to add a write(*,*) here and there, unfortunately passed the 80th column the line will be cut and printed on the following line. Fortunately by just adding these oneliner in the shell before running the program
export FORT_FMT_RECL=200
Will do the trick.
This trick is mainly useful while debugging, when you are writing a program, you should prefer to define your data format as precisely as possible, since you never know how your program will be run or compile.
Hash function race !
 I spent lately a lot of time optimizing my evaluation software. By doing so, I note that with increasing amount of data, time spent in hashing is becoming very long.
I spent lately a lot of time optimizing my evaluation software. By doing so, I note that with increasing amount of data, time spent in hashing is becoming very long.
Thus I make a turn around internet to see whether or not I could optimize my hash function, and finally end up with several good resources among which this website : http://burtleburtle.net/bob/hash/index.html
My problem was that the hash function proposed were in c (and in a marvelous world, I ‘d prefer a Fortran version….that I didn’t find anywhere on the web….so I decided to write it myself.)
I took a list of 235930 pairs of integer (some that are hashed in real evaluations). For each pair, I compute a hash value both with a prime number hash function and with Jenkins’ lookup3.c function.
Technical details :
The prime number hashing function taken as a reference was the one provided by I. Misztal (et al.) in its blupf90 package. Basically, each coordinates to be hashed is multiplied by a specific prime number. The function keep the modulo of the figure obtained by the size of the storage array, where hash key are stored. If for a specific hash key, the cell is already used, then we look for the cell that is 17 cells forward.
In Jenkins’ function, we make several bit operations on the coordinates that we want to hash. In case of collision, we hash again our coordinates until we found an available cell. One important thing is that the storage array size should be a power of two.
Let’s look at the results :
Following are the total number of collisions and the maximum number of collisions for one pair of integer.
 Basically Jenkins hash reduces by a factor 4 the total number of collisions. Furthermore, for a matrix that should be filled at 90% Jenkins’ function won’t have more than 413 successive collisions for one single pair of coordinates, whereas this value could be as high as 8213 with the prime function.
Basically Jenkins hash reduces by a factor 4 the total number of collisions. Furthermore, for a matrix that should be filled at 90% Jenkins’ function won’t have more than 413 successive collisions for one single pair of coordinates, whereas this value could be as high as 8213 with the prime function.
If we observe the evolution of this figure with an increasing proportion of filled cell in our vector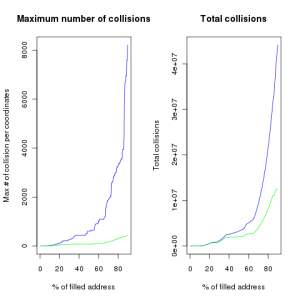 We clearly see that until 40% of the cell are still available, prime hash function tend to generate a lot of collisions, while Jenkins hash remain pretty efficient up to 70% of filled cells.
We clearly see that until 40% of the cell are still available, prime hash function tend to generate a lot of collisions, while Jenkins hash remain pretty efficient up to 70% of filled cells.
Due to the lower level of collisions, Jenkins Hash should be more efficient than Prime function. To test this hypothesis, I then compare the cpu time needed for each algorithm, with or without -03 optimization options with both ifort and gfortran. The test has been repeated 100 times.

 Apart when compiled with gfortran and no options, Jenkins’ hash was significantly faster than the prime function. The difference doesn’t sound huge (about 0.04 CPU s), but should be worthwhile with bigger problem. We can furthermore assume that in real use, we should also get some gains while “looking up” data from the hash matrix.
Apart when compiled with gfortran and no options, Jenkins’ hash was significantly faster than the prime function. The difference doesn’t sound huge (about 0.04 CPU s), but should be worthwhile with bigger problem. We can furthermore assume that in real use, we should also get some gains while “looking up” data from the hash matrix.
For interested people, you can find here a module which can replace the hashvr function in Misztal program.
Beware, you should also :
- add “use hash” in all the subroutine who need hashvr (These subroutine are in sparse.f90)
- Modify the increment size for the hash matrix “hash_incr”(it should be 2)
- Change the default hash size, it should be a power of two.
- Comment the old hashvr function in sparse2.f (to avoid name conflict)
- Update your makefile !
Last hint, if you have a rough idea of the hash matrix size you need, let’s say “H”, you can compute the closest power of two with this code
default_hash_size=2**(1+(log(H)+log10(H))))
You could thus avoid the cost of successive copies of your hash matrix.
